DNA Nanomachines
We
successfully construct
and study two types
of DNA-motors.
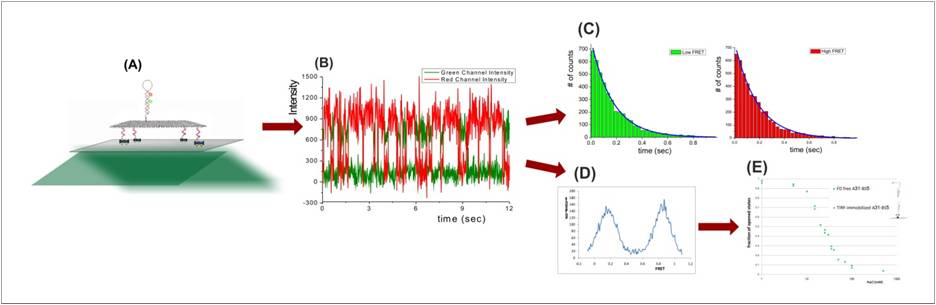
Autonomous motor: We constructed a bipedal autonomous DNA-motor with a coordinate activity between the two motor legs and monitored its activity using SMF techniques. The measurements are done in-situ which enables monitoring the motor’s progress and structural dynamics without disturbing its activity. Our kinetic measurements of the motor’s assembly and activity indicate that it takes dozens of seconds to complete reactions, rather than hours, if components are properly designed. However, we monitor side reactions that significantly reduce the yield of the reaction and resulting in defected motors. We are now working on implementing new strategies for motors’ preparation which will prevent side reactions, altogether, resulting in much higher yield. On the methodological side, we have measured the motor’s dynamics and its interaction with its energy source, a DNA-fuel, in equilibrium and non-equilibrium conditions. Our work demonstrates that by using SMF, one can construct a DNA-machine and monitor its activity in ways not possible with conventional methods. We demonstrate that our methods enable simultaneous in-situ monitoring of the motors efficiency, integrity and activity.
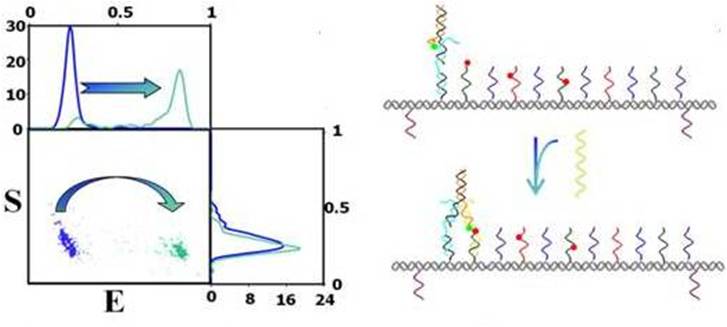
Non-autonomous motor: We constructed a two-leg motor which walks on a DNA-origami track and use energy in the form of fuel/anti-fuel ss-DNAs. The motor successfully completes more than 15 steps and we are now working on increasing the stepping efficiency, speed and walking range. Kinetic measurements indicate that it is possible to significantly improve this kind of motors, such that they will have far higher efficiency and speed. The motor will be capable of maneuvering molecules of interest, e.g. nanoparticles, to a specific location and orientation. Later on, we will use these motors in a device which will maneuver nano-particles in respect to each other in electro-optic device, and a device that exert force on foreign molecules.
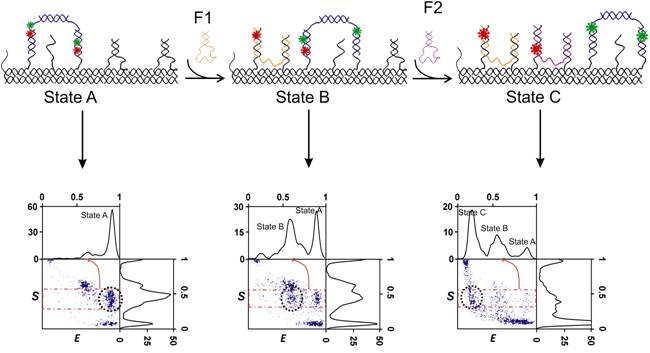
Three
steps detected for Autonomous motor using single-molecule
FRET-ALEX
DNA Dynamics
It is now
understood that
to enable rational
design of nanodevice it is necessary to have a good insight of the
intrinsic DNA
dynamics. For that reason we implement SMF techniques to study the
fundamental
physical rules that govern the behavior of DNA, especially of ss-DNA
hairpin
dynamics. The structural dynamics of hairpins containing various loop
and stamp
sequences are studied when they freely-diffusing and when immobilized
to a
surface. Together, these two complementary methods enable measuring
hairpin
dynamics with a typical time spanning over several orders of magnitude
(dozens
of microsecond to many minutes). For the first time, the hairpins
opening and
closing rates are accurately and directly measured. We are now drawing
general
conclusions regarding ss-DNA dynamics which will enable rational design
of the DNA machines’
moving parts and of fuels.
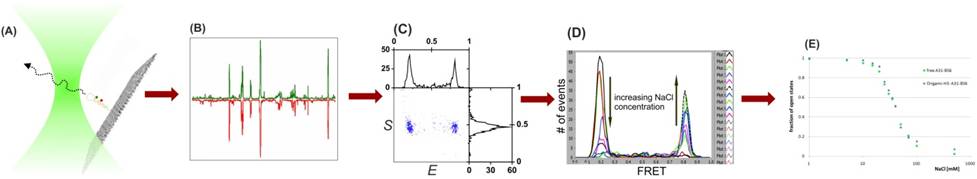

Freely
diffusing approach applied to DNA hairpin
Immobilized approach using TIRF applied to DNA hairpin

Immobilized approach using TIRF applied to DNA hairpin
DNA-Nanoparticles 3D Electronic Devices
We
are using the
specificity and orthogonality
of DNA molecules and DNA-origami to assemble nanoparticles of versions
types in
a particular position and orientation in respect to each other in ways
not
possible with any other method. As a first step, we successfully
position gold
nanoparticles in a specific location on a DNA-origami. We are now
working on
simultaneously positioning several gold nanoparticles and nanoroads on
a single
DNA-origami at specific sites. In collaboration with Taleb
Mokari’s group, we are
planning to position semi-conducting
nanoparticles in
various configurations and study how the nanoparticles relative
configurations
influence the device’s electro-optical properties. Especially
we
are interested
in Plasmon fluoresces enhancement, a promising strategy for efficient
light
harvesting.
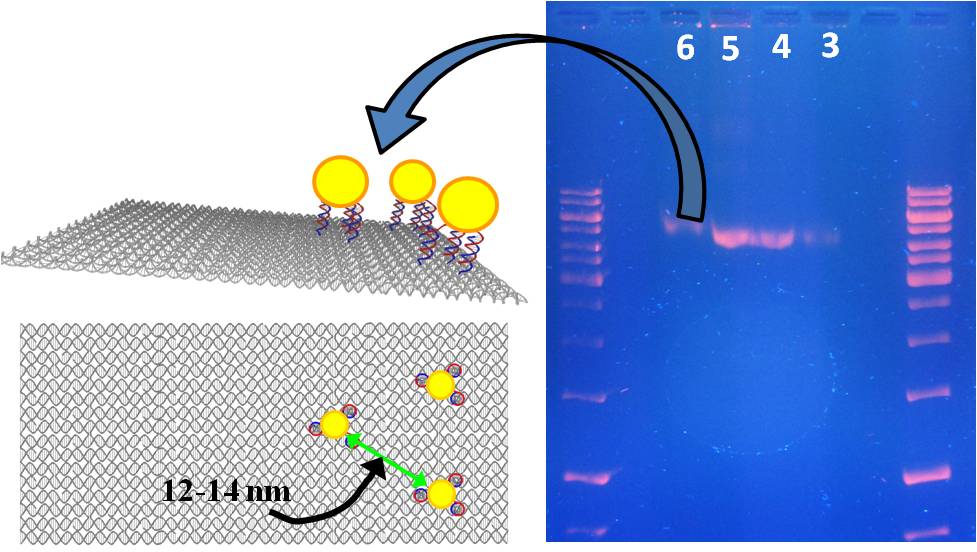
-
Nucleosome Core Particles (NCP)
Nucleosome Core Particles (NCP) are responsible for tightly packing chromosomal DNA and they form an obstacle for regulatory proteins, polymerases, repair and remodeling proteins, all of which require access to DNA for their functionality. The local mechanical properties of DNA, believed to be sequence dependent, are known to play a significant role in formation of a stable NCP. Thus, a good understanding of DNA-related processes and their regulatory functions must include the understanding of affinities between the various nucleosome components, NCP association/dissociation mechanisms and NCP dynamics, and DNA interaction with DNA-binding-proteins, all of the above, in relation to DNA sequence.
Due
to NCP heterogeneous, complexity and dynamic
nature it is adequate to be studied by single-molecule
fluorescence techniques, which enable carful in-situ
structural-dynamics
and interaction investigation of DNA and proteins.
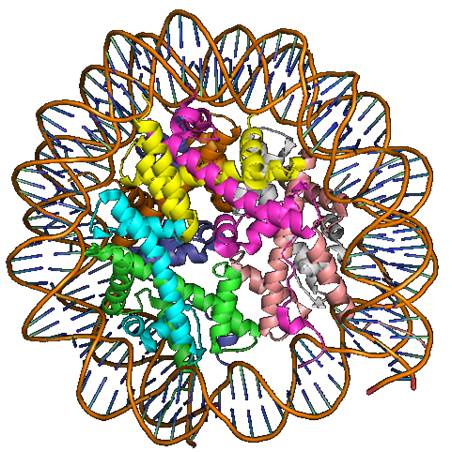
Methodological Development
Our group
specializes in
developing SMF
spectroscopical techniques. We are currently working on several
methodological developments
which will significantly improve the SMF resolution, accuracy and
stability.
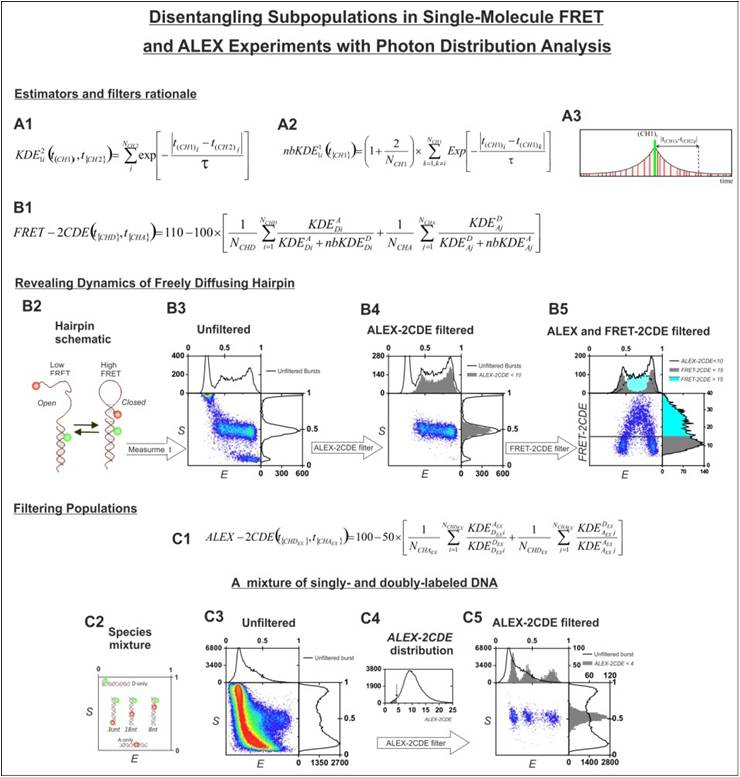
Photons
distribution
analysis: In a
single-molecule
fluorescence measurement of freely diffusing samples, photons belonging
to a
single-molecule (‘burst’) are analyzed in terms of
their
number and not
distribution. As a result, static and dynamic heterogeneity are not
distinguishable, and often, data is wrongly interpreted. We developed a
mathematical algorithm that analyzes the photon distribution inside the
burst,
indicating whether a single-molecule event undergoes dynamics or it is
purely static.
Using our method, it is now possible to analyze complicated dynamical
behavior
in ways not possible with the conventional method. We believe our
straightforward, robust and informative approach will be adapted and
widely used
for analyzing SMF data.
Kinetic measurements: We develop several methods to measure reaction kinetic profiles at the single-molecule level and successfully implement them to monitor DNA-motors assembly and activity reactions (Fig. 2 and Fig 4). Analysis of the kinetic profiles reveals the underlying mechanisms that operate in the motors’ assembling process and in their reaction to fuel and anti-fuel during regular activity. As far as we know, this is a new type of measurements, never done before at single-molecule level and for DNA-motors.