SPdbV provides many different ways to color a model. Color is not a trivial matter in molecular modeling. Colors can reveal structural and chemical features vividly, and can help you to keep your bearings during complex operations.
Select, display, and center the complete model, without side chains. Turn off the display of ribbons, H-bonds, or any other features except the wireframe backbone.
Color: Secondary Structure
SPdbV colors helical residues red, beta sheet
residues (strands) yellow, and all others gray. Notice that these
colors are displayed in small squares beside each residue in the
right-hand column of the Control Panel. Notice also that Color
menu commands colors all groups, regardless of what is
selected.
Color: Secondary Structure Succession
SPdbV colors helices and strands, but with this command, color
reflects the order of each structural element in the overall sequence
of residues. SPdbV colors the first element of secondary structure
violet, the last one red, and the ones in between with colors of the
visible spectrum that lie between violet (400 nm) and red (700 nm).
The result is that it is easy to follow the chain through the protein
-- elements of secondary structure are colored from the N-terminal to
the C-terminal end in the order violet, blue, green, yellow, orange,
red. SPdbV assigns as many shades as needed for the number of
secondary-structural elements present. As before, SPdbV assigns gray
to all other residues.
Select: Nonpolar aa
heading: side
This action adds side chains to selected (in this case, nonpolar)
residues only.
Now notice the letter BS beside the color heading of the Control Panel. Place the pointer on the small black triangle below the color heading and hold down the mouse button. This little menu (the Control Panel color menu) determines whether color commands affect the backbone, sidechains, both, or ribbons. Select sidechain from this menu. Note that the letter S now appears next to color. Subsequent color commands will only affect sidechains.
Color: Type
This command recolors all residues according to
chemical type. Note that it colored only the sidechains because the
Control Panel color menu is set to sidechains only; mainchain
residues remain colored by secondary structure. Nonpolar sidechains
are now gray. Look at the control panel to see the colors assigned to
other types of residues. What are the colors for basic (positive),
acidic (negative), and polar residues? Now look at the model. Notice
that most of the gray side chains on display cluster in the heart of
the molecule, as you would expect, because most hydrophobic side
chains in a water-soluble protein are buried. (Other side chains are
not shown because you selected and displayed nonpolars
only.)
Select: Acidic aa
<control> Select: Basic aa
On the second command, hold down the control
key while pulling down the menu. Using control with a
Select menu command adds groups to the selection, without
un-selecting others.
heading: side
This adds side chains for selected groups, and
removes all other side chains. Look around the model. Most of the red
and blue side chains shown are on the surface, as you should expect
for charged side chains in a water-soluble protein.
Again select, display, and center the complete model, with side chains. Set the Control Panel color menu back to backbone+side.
Color: Accessibility
This operation may take 10 or more seconds, depending
on the speed of your computer. (During slow operations, SPdbV shows a
progress bar to let you know everything is all right.) After the
calculation, you see the model in colors ranging from violet to
orange. The color of each residue is based on the percentage of its
surface area that is exposed (accessible) to the surrounding solvent.
The least accessible (buried) residues are colored violet. To
residues of higher accessibility, SPdbV assigns colors of higher
wavelength in the visible spectrum (the color violet is about 400 nm,
and red is about 700 nm). So in this display, the colors of the
rainbow reflect the exposure of the residue to solvent: violet
residues are the least exposed, red residues the most exposed. Find
tri-NAG (showing surface is a good way to find a group quickly).
Which NAG residues are more accessible?
[Technical note: To be more precise, the accessibility of each residue is computed as the ratio of exposed surface area to the maximum possible exposed surface area. The maximum for residue X is defined as the exposed surface area of residue X in the pentapeptide gly-gly-X-gly-gly in fully extended conformation. To make this calculation, SPdbV adds 1.4 angstroms (approximate radius of a water molecule) to the radius of every atom, computes their surfaces, eliminates all overlapping surfaces, computes the exposed surface area for each residue, computes the ratio of that surface to the maximum possible, and assigns colors on this scale: 75% of maximum exposed: red; 37.5% exposed: green; 0% exposed: violet, with intermediate colors for intermediate values. You can see why even SPdbV, which is a very fast program, takes a few seconds to carry out this calculation.]
Select and center TRP62. With only TRP62 selected and the whole model shown, click on the surface header (small cluster of dots, with a v -- for van der Waals -- at its lower right) in the Control Panel. TRP62 is now shown with a dotted surface that represents the van der Waals radius of each atom. Zoom in so that you can see this residue and its surroundings clearly.
Notice that, just beneath the surface header, there is a small black triangle. This is the handle for another pull-down menu. Click on the triangle to pull the menu down, and select accessible. Notice that the v becomes an a -- for accessible. Again, with the whole model on display, but only TRP62 selected, click on the surface header. A small region of surface dots appears, outside the van der Waals surface. This surface is that portion of the surface of TRP62 that is solvent-accessible, or exposed to the surrounding medium or solvent. You can see that only a small fraction of the TRP62 surface is solvent-accessible, in keeping with the violet color assigned when you colored it by accessibility.
The van der Waals surface lies at a distance of the van der Waals radius from the center of the atom directly below its surface, and includes the full surface of all selected atoms, except where their surfaces overlap. In contrast, the solvent accessible surface lies above each atom at a distance of the van der Waals radius plus 1.4 angstroms, and includes only the surface with which a spherical solvent molecule of radius 1.4 angstroms could come into contact. Any other part of the surface is hidden from the solvent by surrounding residues. Like coloring by accessibility, showing the accessible surface requires a lengthy calculation, and can take a very long time for the entire molecular surface.
Turn off both the van der Waals and accessible surfaces. Center the model, still colored by accessibility.
Display: Slab
This command reduces the display to a thin slab
centered at the middle of the molecule. With this display of the
molecule in cross section, you can see that residues in the heart of
the molecule are not accessible to solvent (dark blue), while surface
residues are accessible. Rotate the molecule to see that this is true
for all the surface.
You can change the thickness of the display slab (to display a thinner or thicker section of the molecule) by clicking on the Screen Attributes button (leftmost icon on the graphics window) and changing the value in the box labeled Slab Depth. You can also move the slab toward or away from you, keeping the same thickness of cross section, by holding down the shift key and dragging the mouse up or down. This works no matter which of the manipulation buttons -- rotate, translate, or zoom -- are currently selected. To return the slab to the center of the model, press the help or = key. Then turn off slabbing by again selecting Display: Slab. Slabs are very handy for eliminating background and foreground groups and exposing the groups you want to study.
The small colored squares in the right-hand column of the Control Panel provide a means to color individual residues, while the word color at the top allows you to choose a color for all selected residues. Display the whole model with side chains, and select all groups except tri-NAG. There are several ways to make this selection. (One quick way is to select NAG 201-203 and then choose Select: Inverse Selection.)
Now change the color of the selected groups. Click to activate the Control Panel.
heading: color
You will see a standard Macintosh color wheel, and
below it, a shade bar. If necessary, slide the pointer on the shade
bar to give the wheel bright colors. Then click in the green region
of the wheel. Click OK. Now the whole model, except for
tri-NAG, which is not selected, is green.
Next, color tri-NAG red, as follows. Select NAG 201-203.
heading: color
Set shade and color to a bright red that will show up
well against the black background. Click OK. Now the green
protein sports a red inhibitor.
You can also color any individual residue by clicking on its individual color box, regardless of whether it is part of the current selection. Try this by coloring LEU129, the C-terminal residue, yellow.
Do you see any other yellow bonds in the model? To help you find them, notice that they disappear when you rotate the model. Identify these yellow structures. Center on one of them as follows:
Click the fourth icon from the right on the graphics window (it has an eye and 4 arrows on it). As instructed below the icons, click an atom -- choose one of the atoms connected by a yellow bond (in stereo, remember to click in the left image). The atoms you chose becomes the center of rotation and display. Zoom in to get a closer look -- slabbing may help.
Select: All
Color: CPK
This action returns all groups to standard or CPK colors: white
for carbon, red for oxygen, blue for nitrogen, and yellow for sulfur.
This color scheme will help you to identify the yellow bonds, which
are common cross-links found mostly in extracellular proteins like
lysozyme. Another way to color selected groups CPK is to click
Cancel on the color wheel dialog.
Test your SPdbV skills:
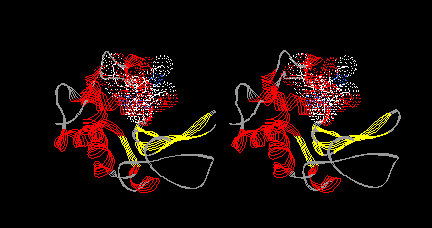
Make a model in which lysozyme is shown only as a ribbon, colored by secondary structure, and tri-NAG is shown in normal (CPK) colors with a dotted van der Waals surface. Once you have it, choose Edit: Reset Orientation and press the help or = key. In stereo, your model should look like this:

Take time to PLAY with the tools introduced in this section.
For more information about coloring, click on SPdbV User Guide in the Contents frame (at left), go to the Display section, and click Color Menu in the third paragraph.